If you are using R to do data analysis inside a company, most of the data you need probably already lives in a database (it’s just a matter of figuring out which one!). However, you will learn how to load data in to a local database in order to demonstrate dplyr’s database tools. At the end, I’ll also give you a few pointers if you do. Syntax การเขียน join ใน R คือ.join(table1, table2, by = 'column') หน้าตาของ dataframe ที่ได้จากการ join แต่ละแบบเหมือนในรูปด้านล่าง โดย ในภาษา R คือค่า missing value (i.e. ไม่สามารถ map ค่าได้จาก. Dplyr is a grammar of data manipulation, providing a consistent set of verbs that help you solve the most common data manipulation challenges: mutate adds new variables that are functions of existing variables. Select picks variables based on their names. Filter picks cases based on their values. Summarise reduces multiple values down to a single summary.
If you are new to dplyr, the best place to start is the data import chapter in R for data science. Installation # The easiest way to get dplyr is to install the whole tidyverse: install.packages('tidyverse') # Alternatively, install just dplyr: install.packages('dplyr') # Or the development version from GitHub: # install.packages('devtools. Lattice R Graphics Cheat Sheet David Gerard 2019-01-22 Abstract: IreproducesomeoftheplotsfromRstudio’sggplot2cheatsheetusingjustthelatticeRpackage.
In a previous post, I described how I was captivated by the virtual landscape imagined by the RStudio education team while looking for resources on the RStudio website. In this post, I’ll take a look atCheatsheets another amazing resource hiding in plain sight.
Apparently, some time ago when I wasn’t paying much attention, cheat sheets evolved from the home made study notes of students with highly refined visual cognitive skills, but a relatively poor grasp of algebra or history or whatever to an essential software learning tool. I don’t know how this happened in general, but master cheat sheet artist Garrett Grolemund has passed along some of the lore of the cheat sheet at RStudio. Garrett writes:
One day I put two and two together and realized that our Winston Chang, who I had known for a couple of years, was the same “W Chang” that made the LaTex cheatsheet that I’d used throughout grad school. It inspired me to do something similarly useful, so I tried my hand at making a cheatsheet for Winston and Joe’s Shiny package. The Shiny cheatsheet ended up being the first of many. A funny thing about the first cheatsheet is that I was working next to Hadley at a co-working space when I made it. In the time it took me to put together the cheatsheet, he wrote the entire first version of the tidyr package from scratch.
It is now hard to imagine getting by without cheat sheets. It seems as if they are becoming expected adjunct to the documentation. But, as Garret explains in the README for the cheat sheets GitHub repository, they are not documentation!
RStudio cheat sheets are not meant to be text or documentation! They are scannable visual aids that use layout and visual mnemonics to help people zoom to the functions they need. … Cheat sheets fall squarely on the human-facing side of software design.
Cheat sheets live in the space where human factors engineering gets a boost from artistic design. If R packages were airplanes then pilots would want cheat sheets to help them master the controls.
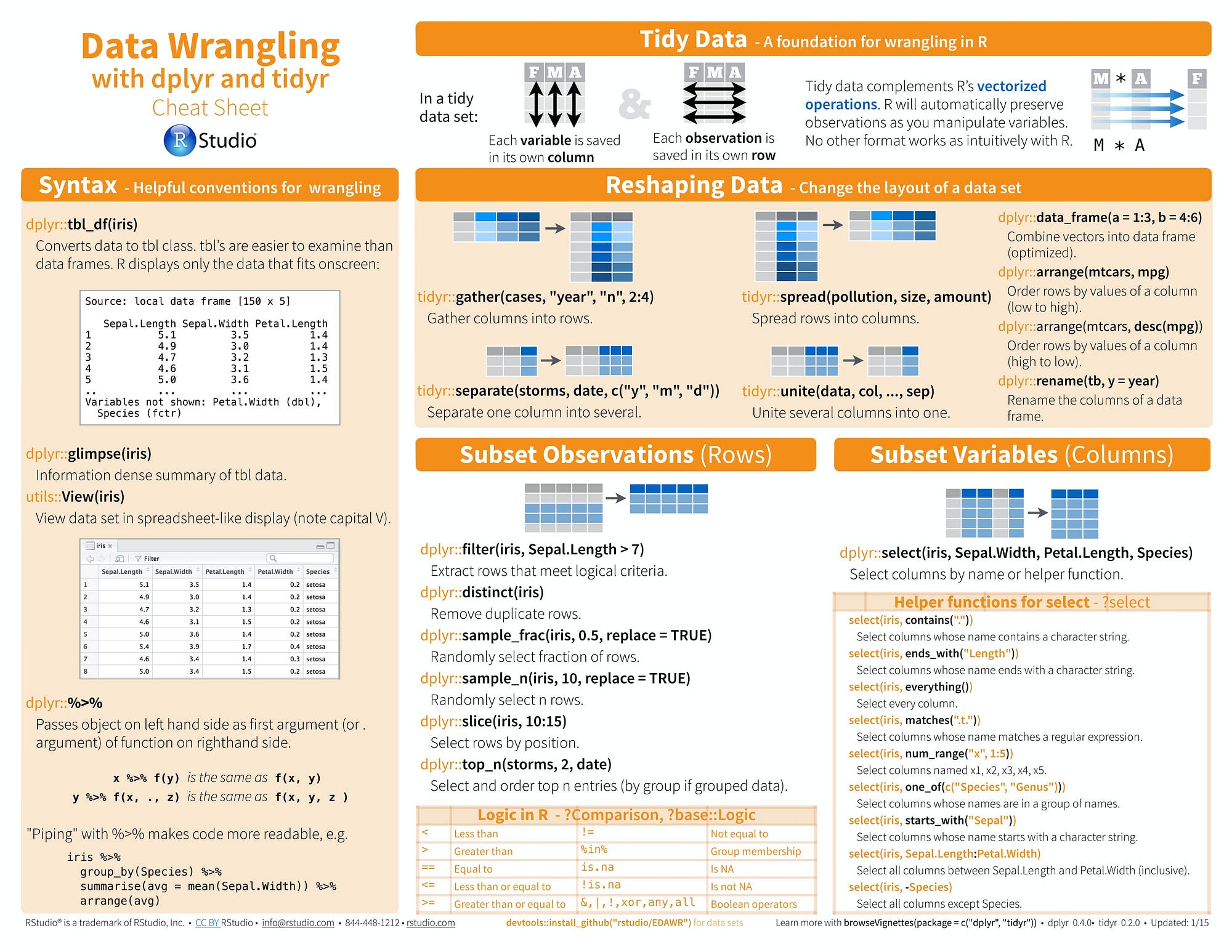
The RStudio site contains sixteen RStudio produced cheat sheets and nearly forty contributed efforts, some of which are displayed in the graphic above. The Data Transformation cheat sheet is a classic example of a straightforward mnemonic tool.It is likely that even someone who just beginning to work with dplyr will immediately grok that it organizes functions that manipulate tidy data. The cognitive load then is to remember how functions are grouped by task. The cheat sheet offers a canonical set of classes: “manipulate cases”, “manipulate variables” etc. to facilitate the process. Users that work with dplyr on a regular basis will probably just need to glance at the cheat sheet after a relatively short time.
The Shiny cheat sheet is little more ambitious. It works on multiple levels and goes beyond categories to also suggest process and workflow.
The Apply functions cheat sheet takes on an even more difficult task. For most of us, internally visualizing multi-level data structures is difficult enough, imaging how data elements flow under transformations is a serious cognitive load. I for one, really appreciate the help.
Cheat sheets are immensely popular. And even in this ebook age where nearly everything you can look at is online, and conference attending digital natives travel light, the cheat sheets as artifacts retain considerable appeal. Not only are they useful tools and geek art (Take a look at cartography) for decorating a workplace, my guess is that they are perceived as runes of power enabling the cognoscenti to grasp essential knowledge and project it in the world.
When in-person conferences resume again, I fully expect the heavy paper copies to disappear soon after we put them out at the RStudio booth.
As well as working with local in-memory data stored in data frames, dplyr also works with remote on-disk data stored in databases. This is particularly useful in two scenarios:
Your data is already in a database.
You have so much data that it does not all fit into memory simultaneouslyand you need to use some external storage engine.
(If your data fits in memory, there is no advantage to putting it in a database; it will only be slower and more frustrating.)
This vignette focuses on the first scenario because it is the most common. If you are using R to do data analysis inside a company, most of the data you need probably already lives in a database (it’s just a matter of figuring out which one!). However, you will learn how to load data in to a local database in order to demonstrate dplyr’s database tools. At the end, I’ll also give you a few pointers if you do need to set up your own database.
Getting started
To use databases with dplyr, you need to first install dbplyr:
You’ll also need to install a DBI backend package. The DBI package provides a common interface that allows dplyr to work with many different databases using the same code. DBI is automatically installed with dbplyr, but you need to install a specific backend for the database that you want to connect to.
Five commonly used backends are:
RMySQLconnects to MySQL and MariaDB
RPostgreSQLconnects to Postgres and Redshift.
RSQLite embeds a SQLite database.
odbc connects to many commercialdatabases via the open database connectivity protocol.
bigrquery connects to Google’sBigQuery.
If the database you need to connect to is not listed here, you’ll need to do some investigation yourself.
In this vignette, we’re going to use the RSQLite backend, which is automatically installed when you install dbplyr. SQLite is a great way to get started with databases because it’s completely embedded inside an R package. Unlike most other systems, you don’t need to set up a separate database server. SQLite is great for demos, but is surprisingly powerful, and with a little practice you can use it to easily work with many gigabytes of data.
Connecting to the database
To work with a database in dplyr, you must first connect to it, using DBI::dbConnect(). We’re not going to go into the details of the DBI package here, but it’s the foundation upon which dbplyr is built. You’ll need to learn more about if you need to do things to the database that are beyond the scope of dplyr.
The arguments to DBI::dbConnect() vary from database to database, but the first argument is always the database backend. It’s RSQLite::SQLite() for RSQLite, RMySQL::MySQL() for RMySQL, RPostgreSQL::PostgreSQL() for RPostgreSQL, odbc::odbc() for odbc, and bigrquery::bigquery() for BigQuery. SQLite only needs one other argument: the path to the database. Here we use the special string, ':memory:', which causes SQLite to make a temporary in-memory database.
Most existing databases don’t live in a file, but instead live on another server. In real life that your code will look more like this:
Rstudio Cheat Sheet Dplyr
(If you’re not using RStudio, you’ll need some other way to securely retrieve your password. You should never record it in your analysis scripts or type it into the console.)
Our temporary database has no data in it, so we’ll start by copying over nycflights13::flights using the convenient copy_to() function. This is a quick and dirty way of getting data into a database and is useful primarily for demos and other small jobs.
As you can see, the copy_to() operation has an additional argument that allows you to supply indexes for the table. Here we set up indexes that will allow us to quickly process the data by day, carrier, plane, and destination. Creating the write indices is key to good database performance, but is unfortunately beyond the scope of this article.
Now that we’ve copied the data, we can use tbl() to take a reference to it:
When you print it out, you’ll notice that it mostly looks like a regular tibble:
The main difference is that you can see that it’s a remote source in a SQLite database.
Generating queries
To interact with a database you usually use SQL, the Structured Query Language. SQL is over 40 years old, and is used by pretty much every database in existence. The goal of dbplyr is to automatically generate SQL for you so that you’re not forced to use it. However, SQL is a very large language, and dbplyr doesn’t do everything. It focuses on SELECT statements, the SQL you write most often as an analyst.
Most of the time you don’t need to know anything about SQL, and you can continue to use the dplyr verbs that you’re already familiar with:
However, in the long run, I highly recommend you at least learn the basics of SQL. It’s a valuable skill for any data scientist, and it will help you debug problems if you run into problems with dplyr’s automatic translation. If you’re completely new to SQL, you might start with this codeacademy tutorial. If you have some familiarity with SQL and you’d like to learn more, I found how indexes work in SQLite and 10 easy steps to a complete understanding of SQL to be particularly helpful.
The most important difference between ordinary data frames and remote database queries is that your R code is translated into SQL and executed in the database, not in R. When working with databases, dplyr tries to be as lazy as possible:
It never pulls data into R unless you explicitly ask for it.
It delays doing any work until the last possible moment: it collects togethereverything you want to do and then sends it to the database in one step.
For example, take the following code:
Surprisingly, this sequence of operations never touches the database. It’s not until you ask for the data (e.g., by printing tailnum_delay) that dplyr generates the SQL and requests the results from the database. Even then it tries to do as little work as possible and only pulls down a few rows.
Dplyr In R Cheat Sheet
Behind the scenes, dplyr is translating your R code into SQL. You can see the SQL it’s generating with show_query():
If you’re familiar with SQL, this probably isn’t exactly what you’d write by hand, but it does the job. You can learn more about the SQL translation in vignette('sql-translation').
Typically, you’ll iterate a few times before you figure out what data you need from the database. Once you’ve figured it out, use collect() to pull all the data down into a local tibble:
collect() requires that database does some work, so it may take a long time to complete. Otherwise, dplyr tries to prevent you from accidentally performing expensive query operations:
Because there’s generally no way to determine how many rows a query willreturn unless you actually run it,
nrow()is alwaysNA.Because you can’t find the last few rows without executing the wholequery, you can’t use
tail().
You can also ask the database how it plans to execute the query with explain(). The output is database-dependent and can be esoteric, but learning a bit about it can be very useful because it helps you understand if the database can execute the query efficiently, or if you need to create new indices.
Creating your own database
If you don’t already have a database, here’s some advice from my experiences setting up and running all of them. SQLite is by far the easiest to get started with, but the lack of window functions makes it limited for data analysis. PostgreSQL is not too much harder to use and has a wide range of built-in functions. In my opinion, you shouldn’t bother with MySQL/MariaDB; it’s a pain to set up, the documentation is sub par, and it’s less feature-rich than Postgres. Google BigQuery might be a good fit if you have very large data, or if you’re willing to pay (a small amount of) money to someone who’ll look after your database.
All of these databases follow a client-server model - a computer that connects to the database and the computer that is running the database (the two may be one and the same, but usually aren’t). Getting one of these databases up and running is beyond the scope of this article, but there are plenty of tutorials available on the web.
MySQL/MariaDB
R Dplyr Cheat Sheet Deutsch
In terms of functionality, MySQL lies somewhere between SQLite and PostgreSQL. It provides a wider range of built-in functions, but it does not support window functions (so you can’t do grouped mutates and filters).
PostgreSQL
PostgreSQL is a considerably more powerful database than SQLite. It has:
a much wider range of built-in functions, and
support for window functions, which allow grouped subset and mutates to work.
BigQuery
BigQuery is a hosted database server provided by Google. To connect, you need to provide your project, dataset and optionally a project for billing (if billing for project isn’t enabled).
It provides a similar set of functions to Postgres and is designed specifically for analytic workflows. Because it’s a hosted solution, there’s no setup involved, but if you have a lot of data, getting it to Google can be an ordeal (especially because upload support from R is not great currently). (If you have lots of data, you can ship hard drives!)
